FLoRIN: Flexible Learning-Free Reconstruction of Imaged Neural Volumes
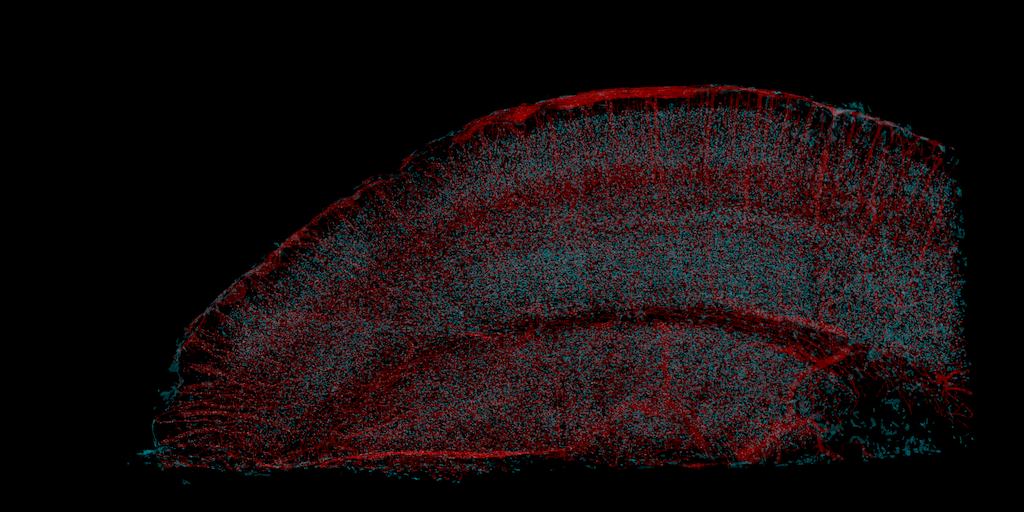
Apex2 Labeled Rodent Brain, Micro-CT X-ray
Imaging technologies in microscopy are the dominant strategy for data collection in neuroscience, able to generate gigabytes to terabytes of data at a time. Historically, segmentation and reconstruction efforts have been carried out manually, but image stacks are becoming prohibitively large. Ideally, a deep learning solution would be able to automatically find microstructures of interest and output reconstructions, however expert annotations are necessary to train learning models.
FLoRIN is a learning-free method for segmentation and reconstruction that meets the needs of neuroscience cross-modality and cross-dataset. Using the novel N-Dimensional Neighborhood Thresholding (NDNT) algorithm and a series of flexible image processing steps, FLoRIN has able to find cells and vasculature in microCT X-ray, filter noise in sSEM to extract APEX2-positive processes, and follow far reaching axons in SCoRe images, among many other applications.
To date, the open source FLoRIN project has collaborated with X labs in Y countries on Z different imaging modalities.
Ali Shahbaz, Jeffery Kinnison, Walter Scheirer
Collaborators: Rafael Vescovi, Ming Du, Robert Hill, Maximillian Jösch, Marc Takeno, Hongkui Zeng, Nunue Maçarico da Costa, Jaime Grutzendler, Narayan Kasthuri
Flexible Learning-Free Segmentation and Reconstruction for Sparse Neuronal Circuit Tracing, Ali Shahbazi, Jeffery Kinnison, Rafael Vescovi, Ming Du, Robert Hill, Maximilian Jösch, Marc Takeno, Hongkui Zeng, Nuno Maçarico da Costa, Jaime Grutzendler, Narayanan Kasthuri, Walter J. Scheirer, March 2018: [pdf]
Reconstruction of Genetically Identified Neurons Imaged by Serial-Section Electron Microscopy, Maximilian Joesch, David Mankus, Masahito Yamagata, Ali Shahbazi, Richard Schalek, Adi Suissa-Peleg, Markus Meister, Jeff W. Lichtman, Walter J. Scheirer, Joshua R. Sanes, eLife, July 2016: [pdf]
[code]